Authors:
Kimberly J. Felcher, Michigan State University; David Douches, Michigan State University
Objectives
To give an example of how a molecular marker can be used in a potato breeding program.
Introduction
Potato breeding programs evaluate and select for numerous traits driven by end-use (fresh-market, chip processing, frozen/french fry processing, novelty), consumer preferences (skin color, flesh color, shape, culinary quality), and production requirements (yield, maturity, disease resistance). For a new cultivar to be successful it must combine as many of the desired traits as possible. For example, a new potato chip cultivar must meet grower demands (high yield, disease resistant, early maturing) and processor demands (high specific gravity, low levels of reducing sugars, smooth/round shape). The selection process occurs over multiple locations; includes field, greenhouse, and laboratory assessments; and requires 10–15 years from the initial cross to the release of a new variety. Furthermore, because trait demands can change over the short-term and the long-term, breeders must maintain and develop a germplasm pool with high frequencies of genes for desirable traits. One way to increase the efficiency of the breeding/selection process is marker-assisted selection (MAS). While many marker/trait association studies have been conducted in potato, only a few of these markers are being used for MAS due to cost, population specificity, linkage distance, and the amount of trait variation the markers account for. One marker with practical application for potato breeding is the RYSC3 marker for potato virus Y (PVY) resistance.
PVY Resistance Genes and Associated Molecular Markers
PVY is a member of the Potyvirus group that can reduce potato yield and quality. It is a particular problem for seed production as it can cause the rejection of seed lots in certification programs. Several genes for PVY resistance have been identified in wild species and introgressed into the cultivated potato. These include: Ryadg from Solanum tuberosum ssp. andigena (Kasai et al., 2000), Rysto from Solanum stoloniferum (Brigneti et al., 1997), and Ry-fsto from Solanum stoloniferum (Flis et al., 2005). The Ryadg gene confers extreme resistance to PVY and co-segregates with the PCR-based, SCAR marker RYSC3 (Kasai et al., 2000). This marker is suitable for MAS for several reasons: it co-segregates with the trait (PVY resistance), it requires the use of very little DNA, it is easily scored (presence/absence of a single band), it takes only a few hours to use, thus many samples can be analyzed per day, and the cost per sample is relatively low. Furthermore, an extensive study by Whitworth et al. (2009) demonstrated that the RYSC3 marker could identify clones with resistance to all PVY strains that are present in North America.
Overview of Potato Breeding and the Use of the RYSC3 Marker for MAS
Potato breeding can best be described as pedigree breeding where phenotypic selection (based primarily on maturity, tuber appearance, and yield) is done at the F1 generation (year 1/single hill selections) and the genotype is then fixed due to clonal propagation. Selection continues over several years to find clones with the desired combination of traits (tuber quality, disease resistance, processing quality, increased nutritional value). These are traits that require larger plot sizes, more replications, and testing in multiple environments for accurate characterization. It isn’t feasible, due to cost and time constraints, to test several thousand, single-hill selections for PVY resistance in inoculated greenhouse studies. A more cost effective option is to make year 1 selections, where > 90% of the progeny are discarded, and then identify PVY-resistant progeny with the RYSC3 marker. However, only clones whose pedigrees indicate the potential for the presence of the Ryadg gene are subjected to MAS using the RYSC3 marker. Because PVY resistance is important but not necessary for the success of a new variety, the presence of the RYSC3 marker does not guarantee the advancement of a clone at this stage. However, the information is documented and may influence the selection process at later generations or the choice of parents to include in future crosses. Those clones that have the RYSC3 marker at the advanced breeding line stage are phenotyped for PVY resistance prior to commercialization.
Genotyping
Template DNA is extracted from leaf tissue that is collected from F1 plants growing in the field or from post-harvest tuber tissue. A high-throughput, 96-well, DNA extraction protocol is used to increase the efficiency and cost effectiveness of the process. Following PCR, using the template DNA and the RYSC3 primers designed by Kasai et al (2000), and electrophoretic separation, the individual clones are scored for the presence or absence of the 321 bp band that is linked to PVY resistance (Ryadg) (Fig. 1).
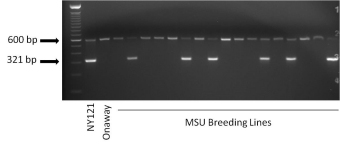
Figure 1. Example of a gel showing the presence or absence of the RYSC3 marker. Lines with a 321 bp band (such as NY121, a positive control known to have the Ryadg gene) are RYSC3 positive and lines lacking the band (such as Onaway, a negative control known to lack the Ryadg gene) are RYSC3-negative. The 600 bp band that is present in all lanes is an internal control used to verify that the PCR reaction worked properly. Photo credit: Dan Zarka, Michigan State University Potato Breeding and Genetics Program.
Phenotyping
Advanced clones that have commercial potential and are RYSC3 positive are evaluated for PVY resistance via manual inoculations of multiple PVY strains (PVY0, PVYNTN, PVYN:0) followed by ELISA testing (Whitworth et al., 2009).
Conclusions
Although MAS is not widely used in potato breeding at present, the RYSC3 marker is a practical example of how MAS can be beneficial for the development of new varieties and enhanced breeding populations. As our knowledge of the potato genome grows and more genes are identified and linked to important traits, MAS will almost certainly become an increasingly important tool used by potato breeders.
References Cited
- Brigneti, G., J. Garcia-Mas, and D. C. Baulcombe. 1997. Molecular mapping of the potato virus Y resistance gene Rysto in potato. Theoretical and Applied Genetics 94: 198–203. (Available online at: http://dx.doi.org/10.1007/s001220050400) (verified 12 May 2012).
- Flis, B., J. Hennig, D. Strzelczyk-Zyta, C. Gebhardt, and W. Marczewski. 2005. The Ry-fsto gene from Solanum stoloniferum for extreme resistance to potato virus Y maps to potato chromosome XII and is diagnosed by PCR marker GP122718 in PVY resistant potato cultivars. Molecular Breeding 15: 95–101. (Available online at: http://dx.doi.org/10.1007/s11032-004-2736-3) (verified 12 May 2012).
- Kasai, K., Y. Morikawa, V. A. Sorri, J.P.T. Valkonen, C. Gebhardt, and K. N. Watanabe. 2000. Development of SCAR markers to the PVY resistance gene Ryadg based on a common feature of plant disease resistance genes. Genome 43: 1–8. (Available online at: http://dx.doi.org/10.1139/gen-43-1-1) (verified 12 May 2012).
- Whitworth, J. L., R. G. Novy, D. G. Hall, J. M. Crosslin, and C. R. Brown. 2009. Characterization of broad spectrum potato virus Y resistance in a Solanum tuberosum ssp. andigena-derived population and select breeding clones using molecular markers, grafting and field inoculations. American Journal of Potato Research 86: 286–296. (Available online at: http://dx.doi.org/10.1007/s12230-009-9082-2) (verified 12 May 2012).
External Links
- Wikipedia contributors. 2010. Marker Assisted Selection [online]. Wikipedia, The Free Encyclopedia. Available at: http://en.wikipedia.org/w/index.php?title=Marker_assisted_selection&oldi… (verified 12 May 2012).
- Wikipedia contributors. 2010. Potato virus Y [online]. Wikipedia, The Free Encyclopedia. Available at: http://en.wikipedia.org/w/index.php?title=Potato_virus_Y&oldid=396564236 (verified 12 May 2012).
- Wikipedia contributors. 2010. ELISA [online]. Wikipedia, The Free Encyclopedia. Available at: http://en.wikipedia.org/w/index.php?title=ELISA&oldid=402585579 (verified 12 May 2012).
Funding Statement
Development of this article was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, agreement 2009-85606-05673, administered by Michigan State University. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author and do not necessarily reflect the view of the United States Department of Agriculture.
PBGworks 882
