Author:
Carlos M. Hernandez-Garcia, The Ohio State University
Introduction
Modern plant breeding relies on the manipulation of desirable characteristics directly determined by genetics. Most of the genes governing discrete characters in plants are regulated at transcriptional levels; however, determination of seed coat color in soybean is a rare example of a post-translationally regulated phenotype. Although genes responsible for seed coat pigmentation –CHS chalcone synthases– are functional and normally transcribed, their mRNAs are targeted for destruction via an RNA-silencing mechanism, therefore leading to colorless or light-yellow seed coats. This mechanism was not well understood until recently (Tuteja et al. 2009). Seed coat pigmentation is widely conserved in wild soybeans and is mainly due to the presence of pigments such as anthocyanins. However, the development of modern varieties has been focused on colorless or light-yellow seed coat phenotypes as pigments can interfere with protein and oil extraction during industrial processing.
In soybean, four naturally occurring alleles (I, ii, ik, i) of the I (inhibitor) locus govern seed coat pigmentation. The inhibitor dominant alleles I, ii, and ik lead to a colorless or light yellow seed coat, a pigmented hilum, and regions of saddle-shaped pigmentation, respectively (Fig. 1). In contrast, the recessive i allele gives rise to pigmented black or brown seed coats (Todd and Vodkin, 1993). Spontaneous mutations of either the I or ii allele can also cause reversion to the pigmented seed coat phenotype. The structure of the I locus has been widely documented (Todd and Vodkin, 1996; Tuteja and Vodkin, 2008) as a region mapping to chromosome 8 that contains two perfectly repeated and inverted clusters of the CHS1, 3, and 4 genes. Six additional CHS genes –CHS2, 5, 6, 7, 8, and 9– are also present in the soybean genome. CHS is the first enzyme in the phenylpropanoid pathway that leads to a diverse range of secondary metabolites including anthocyanin pigments present in various plant tissues, such as flowers and seed coats.

Figure 1. Seed coat phenotypes governed by four alleles of the soybean I locus. Note that the recesive i alleles can lead to both black and brown seed coats. Figure credit: Carlos M. Hernandez-Garcia, The Ohio State University.
Previous investigations conducted by Tuteja et al. (2004) revealed that CHS gene transcripts, particularly the nearly identical CHS7-8 transcripts, which are responsible for seed coat pigmentation, are down regulated in cultivars with non-pigmented seed coat phenotypes carrying either the I or ii locus. However, CHS7-8 transcripts are paradoxically abundant in pigmented seed coats of cultivars carrying the mutated I-locus, demonstrating that this locus is directly responsible for suppression of pigmentation. Moreover, CHS genes are expressed in other plant tissues, suggesting a highly tissue-specific inhibition of CHSs exclusive to seed coats.
With the advent of next generation sequencing, Tuteja et al. (2009) demonstrated that abundant CHS small interfering RNAs (siRNAs), predominantly of 22 nucleotides, were produced as a result of the I locus in non-pigmented soybeans. These siRNAs were specific to seed coats and almost undetectable in developing cotyledons and other tissues. Conversely, a very low abundance of CHS siRNAs was detected in pigmented-seed coats from a cultivar containing the homozygous recessive i allele. Tuteja et al. (2009) showed evidence suggesting that transcription of CHS1, 3, and 4 arranged in the inverted repeats is performed normally, but results in a double-stranded RNA (dsRNA), which is further processed into primary siRNAs by DICER enzymes. Evidence also suggests that the resulting primary CHS1, 3, and 4 siRNAs are assembled with mature CHS7-8 mRNAs in an RNA-dependant RNA polymerase complex (RdRP) to produce secondary siRNAs capable of amplifying the RNA silencing signal and targeting mRNAs of all 9 CHSs for degradation (Fig. 2).
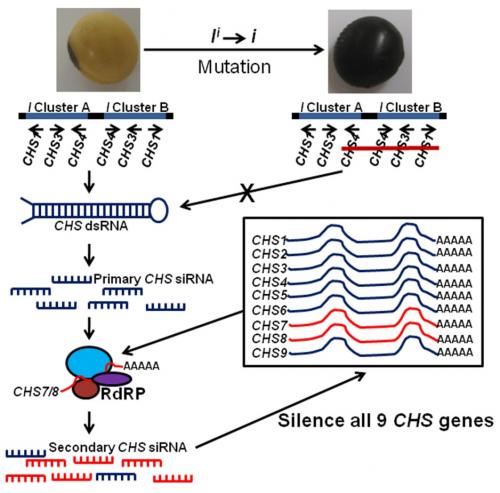
Figure 2. RNA silencing of CHS (chalcone synthase) genes in soybean seed coats due to the inverted repeats containing the CHS1, 3, and 4 genes. Figure adapted from Tuteja et al. (2009).
To date, very few examples of traits being regulated by an RNA-silencing mechanism have been reported in plants. The tissue-specificity of the CHS siRNA biogenesis is intriguing and remains to be deciphered. Global RNA sequencing as performed in this study for siRNA is a powerful tool to study transcript profiling, and may be fundamental to the discovery of other traits regulated by RNA silencing.
References Cited
- Iezzoni, A. 2010. Jewels in the Genome. RosBREED Newsletter 1(1): 9. (Available online at: http://rosbreed.s3.amazonaws.com/wp-content/uploads/2010/10/2010-02_RosBREED.Newsletter.pdf) (verified 19 Apr 2011).
- Todd, J. J., and L. O. Vodkin. 1993. Pigmented soybean (Glycine max) seed coats accumulate proanthocyanidins during development. Plant Physiology 102:663-670.
- Todd, J. J., and L. O. Vodkin. 1996. Duplications that suppress and deletions that restore expression from a chalcone synthase multigene family. Plant Cell 8:687-699.
- Tuteja, J. H., and L. O. Vodkin. 2008. Structural features of the endogenous CHS silencing and target loci in the soybean genome. Crop Science 48:49-69. (Available online at: http://dx.doi.org/10.2135/cropsci2007.10.0542tpg) (verified 8 July 2011).
- Tuteja, J. H., S. J. Clough, W. C. Chan, and L. O. Vodkin. 2004. Tissue-specific gene silencing mediated by a naturally occurring chalcone synthase gene cluster in Glycine max. Plant Cell 16:819-835. (Available online at: http://dx.doi.org/10.1105/tpc.021352) (verified 8 July 2011).
- Tuteja JH, Zabala G, Varala K, Hudson M, Vodkin LO (2009) Endogenous, tissue-specific short interfering RNAs silence the chalcone synthase gene family in Glycine max seed coats. Plant Cell 21:3063-3077. (Available online at: http://dx.doi.org/10.1105/tpc.109.069856) (verified 8 July 2011).
Acknowledgments
The author would like to thank Drs. David Francis and Heather Merk for critically reading this review and providing helpful suggestions. The author also thanks Sungwoo Lee for providing soybean seeds depicted in figures 1 and 2.
Funding Statement
Development of this lesson was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, agreement 2009-85606-05673, administered by Michigan State University and CONACYT, Mexico. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the United States Department of Agriculture or any of the other aforementioned entities.
PBGworks 1093
