Author:
Josh Clevenger, The Ohio State University
Introduction
Wild relatives of cultivated tomato bear fruit that can be several hundred times smaller than cultivated varieties (Solanum lycopersicum). Additionally, cultivated fruit varieties contain more locules than their wild progenitors. Tomato fruit weight is controlled by a few quantitative trait loci (QTL), one of which, fw2.2, results in a regulatory change in a cell-division gene (Frary et al. 2000). The extreme size of some cultivated varieties, although, cannot be explained by the fw2.2 QTL alone, as all cultivated varieties contain the large-fruited allele. The jump from medium-sized to larger-sized fruit in high locule number varieties has been attributed to two QTL that control locule number on chromosome 2 (locule number (lc)) and chromosome 11 (fasciated (fas)).

Figure 1. Difference in size between a fruit with many locules (LA352) with the fas mutant allele and a fruit with two locules (LA3263) with the wild type FAS allele. Image credit: Josh Clevenger, The Ohio State University.
Locule Number
The number of locules a tomato fruit develops is determined by the number of carpels in the flower. Wild species of tomato have fruit with 2-4 locules, while cultivated varieties containing the mutant fas allele can develop more than 15 locules. This can increase fruit size by as much as 50% (Lippman and Tanksley, 2001). Thus, the increase in locule number is an important step in the development of larger tomato fruit.
Molecular Basis of fas
The underlying molecular basis for fas has been established by Steven Tanksley’s group at Cornell (Cong et al. 2008). QTL studies have shown that the fas locus resides on the long arm of chromosome 11 (Cong et al. 2008). This locus is believed to be the same one described by MacArther (f) (1934). In order to fine map the locus, a cross was made between LA2371, a tomato cultivar with high locule number, and an introgression line containing the low locule fas allele from LA716 (S. pennellii). The F2 population was then screened for recombination events between two markers that flank the fas locus, T302 and cLET24J2A (Table 1). Thirty plants were selected to derive F3 progeny for further marker evaluation. The results of the progeny test indicated that fas cosegregates with the markers t1948 and TG105A (Cong et al. 2008).
Table 1. Summary of the markers used to fine map the fas locus. Modified from Cong et al. 2008.
| Marker | Forward Primer (5′-3′) | Reverse Primer (5′-3′) | Marker Type | Restriction Enzyme | Source |
|---|---|---|---|---|---|
| T302 | TGGCTCATCCTGAAGCTGATAGCGC’ | AGTGTACATCCTTGCCATTGACT | INDEL/RFLP | XbaI | Clone cLED24J21 |
| cLET24J2A | CAACCATCCTAGCAATGAAATCT | GAGGCATTCACTCTCTTCGATAC | CAP/RFLP | KpnI/ScaI | cDNA clone |
| t1948 | TTCCCAATTCACCAATTTTTC | ATCCAGAACCAGCACATTTCTT | CAP/RFLP | DraI/EcoRI/V | Clone cLET45E9 |
| TG105A | N/A | N/A | RFLP | DraI | Genomic Clone |
Two overlapping BAC clones were isolated that contain the region between T1948 and TG105A and the subsequent contig was sequenced and annotated for genes. The contig contained 39 putative genes. A homologous function analysis led to the identification of two likely candidate genes, a YABBY-like protein and a TOUSLED-like protein (Cong et al. 2008). YABBY, a transcription factor, regulates flower organ number in rice (Jang et al. 2004). TOUSLED, a protein kinase, controls floral organ development (Roe et al. 1993). After analyzing the transcription levels and ascertaining sequence data from these two candidate proteins, YABBY-like showed significant differences in expression and sequence between the wildtype and mutant populations (Cong et al. 2008).
To test whether the YABBY-like protein was in fact fas, the gene plus upstream and downstream coding regions from a low locule variety (TA492, a two locule S. lycopersicon accession) was transformed into a high locule variety (LA2371). All transformants contained a significant decrease in locule number. From this data the authors concluded that fas encodes a YABBY-like transcription factor that regulates locule number in tomato (Cong et al. 2008).
To further elucidate the molecular basis for fas, expression and sequence analysis were performed. Authors reported that during flower development, fas is down regulated in high locule number varieties (Cong et al. 2008). Sequence analysis of high and low locule number varieties showed a large insertion in the first intron of all high locule number varieties tested and an absence of the insertion in all low locule number varieties tested (Fig. 2). It was concluded that the high locule number phenotype caused by fas is due to a down regulation of the YABBY-like protein caused by a large insertion in the first intron (Cong et al. 2008).
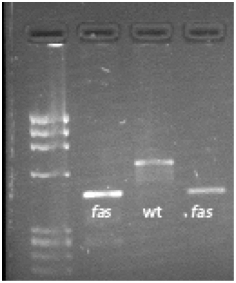
Figure 2. PCR detection of fas inserion. From left to right; many loculed variety Orange Strawberry, two locule wild species LA1589, and many loculed variety LA352. Primers used in a 3 primer reaction: 5’ – CCAATGATAATTAAGATATTGTGACG – 3’, 5’ – CAGAAATCAGAGTCCAATTCCA – 3’, 5’ – GTCACTCTGTCTTGTTACTGC – 3’. Image credit: Zejun Huang and Josh Clevenger, The Ohio State University.
Conclusion
The fas mutation is an excellent example of a trait that can be efficaciously selected for using a molecular marker. Individuals homozygous for fas have up to 15 or more fruit locules and fruit can be 50% larger than individuals that do not contain the mutation. A molecular marker for fas (Fig. 2), along with other fruit weight QTL can be used early in a breeding program and select immediately for larger tomatoes, allowing the breeder more time to concentrate phenotyping efforts on other desirable traits.
References Cited
- Cong B., L. S. Barrero, and S. D. Tanksley. 2008. Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nature Genetics 4: 800-804. (Available online at: http://dx.doi.org/10.1038/ng.144) (verified 21 July 2011).
- Frary, A., T. C. Nesbitt, S. Grandillo, E. van der Knaap, B. Cong, J. P. Liu, J. Meller, R. Elber, K. B. Alpert, and S. D. Tanksley. 2000. fw2.2: a quantitative trait locus key to the evolution of tomato fruit size. Science 289: 85–88. (Available online at: http://dx.doi.org/10.1126/science.289.5476.85) (verified 21 July 2011).
- Iezzoni, A. 2010. Jewels in the Genome. RosBREED Newsletter 1(1): 9. (Available online at: http://rosbreed.s3.amazonaws.com/wp-content/uploads/2010/10/2010-02_RosBREED.Newsletter.pdf) (verified 19 Apr 2011).
- Jang, S., J. Hur, S. J. Kim, M. J. Han, S. R. Kim, and G. An. 2004. Ectopic expression of OsYAB1 causes extra stamens and carpels in rice. Plant Molecular Biology 56: 133-143. (Available online at: http://dx.doi.org/10.1007/s11103-004-2648-y) (verified 21 July 2011).
- Lippman, Z., and S. D. Tanksley. 2001. Dissecting the genetic pathway to extreme fruit size in tomato using a cross between the small-fruited wild species Lycopersicon pimpinellifolium and L. esculentum var. Giant Heirloom. Genetics 158: 413–422.
- Macarthur, J. W. 1934. Fruit size effects of qualitative genes in the tomato. American Naturalist 58:73-74.
- Roe, J. L., C. J. Rivin, R. A. Sessions, K. A. Feldmann, and P. C. Zambryski. 1993. The TOUSLED gene in A. thaliana encodes a protein kinase homolog that is required for leaf and flower development. Cell 75: 939–950.
Funding Statement
Development of this page was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, agreement 2009-85606-05673, administered by Michigan State University. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the United States Department of Agriculture.
PBGworks 1094
