Author:
Shaun Broderick, The Ohio State University
Introduction
The floricultural market is highly competitive, and floricultural crops vary widely in popularity from one year to the next. Consumer demand of traditional and (or) commonplace bedding plants, such as petunia, is often driven by novel and unusual characteristics. The cultivated Petunia x hybrida is thought to have originated from a cross between P. axillaris and P. integrifolia made nearly two centuries ago (Armitage, 2001). Since those original crosses and subsequent breeding programs, the color palette has been widely expanded and flower morphologies have been altered drastically.
Bi-colored flowers have 3 standard coloration patterns: Picotee flowers have colored centers and white margins, Morn flowers have a white center and colored margins, and Star flowers have five radiating stripes of contrasting color (Matsubara et al. 2008; Fig. 1). With such a rich array of petunia phenotypes already available, truly novel traits are rare.

Figure 1. Bi-colored petunia flower patterns. A) Picotee (Dreams Red Picotee), B) Star (Madness™ Burgundy Star), C) Morn (Mirage Rose Morn), and D) putative fbp2 phenotype (Pretty Much Picasso™, ‘BTUN31501’). Image credits: a-c) Ball Horticultural Company d) Shaun Broderick, The Ohio State University.
The Novel Picotee Phenotype
In 2007, the petunia color palette was further expanded with a new Picotee patterned variety, ‘Pretty Much Picasso™ ‘BTUN31501’, which develops a green margin (Fig. 1). Although similar phenotypes have been previously reported (Agenent et al. 1994; Vandenbussche et al. 2003; and Bailey, 1896), ‘BTUN31501’ is the first commercially available petunia of its type. Matsubara et al. (2008) studied the genetic components of this phenotype using a wild Paraguayan petunia population. Their search revealed a mutant floral binding protein 2 (fbp2) was the cause of this novel phenotype. An overview on flower development aids in understanding the mechanism through which fbp2 affects flower morphology and color.
Floral Development
Beginning in the center of the flower and moving outward, all complete flowers consist of four whorls: carpels, stamens, petals, and sepals. This order is established very early within flower development. Coen and Meyerowitz (1991) outlined the current ABC model of flower development, which was formulated by discovering specific homeotic mutant lines of Arabidopsis thaliana and Antirrhinum majus. Homeotic genes are responsible for whorl organization during development. By finding individuals with mutant homeotic flower phenotypes, researchers were able to determine the genes involved in flower formation.
Genes specific to each flower part are controlled by a specific combination of three transcription factor classes: A, B, and C. Each of the genes belonging to these classes contain a MADS-box DNA binding domain which recognizes and binds to specific homeotic genes. Within the meristem of a primordial flower, classes A and C are expressed in adjacent cells, while expression of class B overlaps part of A and C. This arrangement allows the formation of four unique combinations of transcription factors: A, AB, BC, and C. Each combination is necessary and specific for the proper formation of their respective whorl. Class A alone regulates the genes necessary for sepal formation, classes A and B together regulate petal formation, classes B and C regulate stamen formation, and class C regulates carpel formation. Mutations in these transcription factors often result in improper flower formation. For example, if transcription factor B has a non-functional mutation, the developing flower would only consist of sepals and carpels because only classes A and C would be present. If transcription factor A was non-functional, the flower would only develop stamens and carpels.
The ABC model was further expanded after the discovery of SEPALLATA1 (SEP1), SEP2, and SEP3 homeotic transcription factors, which precede classes B and C in Arabidopsis whorl development. Pelaz et al. (2000) demonstrated that the SEP gene family was necessary for B and C class transcription factors to fully develop. In triple mutants containing sep1, sep2, and sep3, the floral tissues such as petals and stamens were replaced by sepal-like tissue. The SEP transcription factors span the petals, androecium, and gynoecium of developing flowers. In work done in petunia, FBP2 was found to be an ortholog to the SEP transcription factors and appear to share similar phenotypes to the Arabidopsis mutants (Angenent et al. 1994).
Genetics of the Green Picotee Petunia
To determine the genetic factors specific to the wild Paraguayan petunia with the green Picotee pattern, Matsubara et al. (2008) isolated and sequenced the Floral Binding Protein 2 (FBP2) (GenBank accession number M91666, Angenent et al. 1992) gene from a wild-type petunia and the mutant P. inflata. Although the exons (coding sequence) were found to be of similar lengths in both the wild-type and mutant petunias, intron (non-coding region) II was found to be much longer (by 2,123 bp) in the mutant P. inflata.
For proper flower development, FBP2 must be transcribed in high enough quantities to override the default developmental pathway of epidermal (leaf) tissue. Matsubara et al. (2008) measured FBP2 expression and found that it was greatly diminished in the mutant petunia. DNA sequencing unveiled a transposon (a DNA segment that can be randomly inserted throughout a genome) called dPifTp1 disrupting intron II (Table 1). In this particular instance, dPifTp1 impacts the normal transcription of FBP2, and the abundance of fbp2 mRNA transcripts was diminished. Although fbp2 transcripts were found within the margins of the corollas, levels in the mutant petunias were found to be much lower than wild type. With these lower levels, the formation of epidermal tissue with stomata and trichomes occurred at the margins of corolla which induce the green margin phenotype.
This trait is inherited in a single-gene homozygous recessive pattern (i.e. 3:1 segregation ratio). When selfed, approximately 25% of progeny display the novel phenotype. Matsubara et al. (2008) designed primers that can be used to distinguish mutant and wild-type FBP2 phenotypes within a breeding program (Table 1; Fig. 2).
| Table 1. FBP2 sequencing primers. | |||
|---|---|---|---|
| Name | Primer sequences | Amplicon description | GenBank accessions |
| fbp2-f1 | 5ʹ-TATGGGAAGAGGTAGAGTTGAGCTT-3ʹ | Full-length genomic(g)DNA or cDNA | gDNA: AB375308, AB375309; cDNA: M91666 |
| fbp2-r5 | 5ʹ-CAACCAGCCAGCCATGTAGTT-3ʹ | ||
| fbp2-f7 | 5ʹ-CTCTAAACCCCTCCCCACC-3ʹ | dPifTp1 | N/A |
| fbp2-r11 | 5ʹ-TGAAATCCCCCAAAGAACAATATT-3ʹ | ||
| fbp2-f3 | 5ʹ-GCACCAGAGACTAAATATCCACACG-3ʹ | Intron 2 of the genomic DNA FBP2 size: 1.7 kbp; fbp2 size: 3.8 kbp | gDNA FBP2: AB375307 |
| fbp2-r3 | 5ʹ-TTGGCTGCTTATTTCCTGTAATCAT-3ʹ |
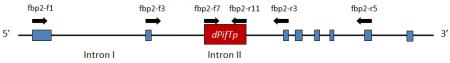
Figure 2. Structural organization of the fbp2-3 allele containing dPifTp1. The exons are represented by blue boxes. Introns are represented by the black line between the blue boxes. The transposon, dPifTp, is not present in the wild-type FBP2. Arrows indicate the approximate annealing position of the respective primers. (Adapted from Matsubara et al. 2008).
Conclusion
Utilization of this novel trait within petunia breeding programs may create novel phenotypic combinations. For example, this trait could be incorporated into elite petunia lines with larger corollas. Also, unique color combinations could be achieved by incorporating this trait into petunia flowers of different colors. Such crosses have the potential to create striking cultivars high consumer demand. The discovery of such a unique trait within a wild population demonstrates the need to maintain and preserve wild species for future breeding programs. Continued evaluation of wild germplasm may reveal more novel phenotypic breakthroughs that could have profound marketing value. If this source for genetic diversity is lost, future breeding efforts may be suppressed.
References Cited
- Angenent, G. C., J. Franken, M. Busscher, D. Weiss, and A. J. Van Tunen. 1994. Co-suppression of the petunia homeotic gene fbp2 affects the identity of the generative meristem. Plant Journal 5:33–44.
- Armitage, A. M. 2001. Armitage’s manual of annuals, biennials, and half-hardy perennials. Timber Press, Portland, OR.
- Bailey, L. H. 1896. Evolution of the Petunia. p. 465-472. In: L. H. Bailey. The survival of the unlike. Macmillan, London.
- Coen, E. S., and E. M. Meyerowitz. 1991. The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37.
- Iezzoni, A. 2010. Jewels in the Genome. RosBREED Newsletter 1(1): 9. (Available online at: http://rosbreed.s3.amazonaws.com/wp-content/uploads/2010/10/2010-02_RosBREED.Newsletter.pdf) (verified 19 Apr 2011).
- Matsubara, K., K. Shimamura, H. Kodama, H. Kokubun, H. Watanabe, I. L. Basualdo, and T. Ando. 2008. Green corolla segments in a wild Petunia species caused by a mutation in FBP2, a SEPALLATA-like MADS box gene. Planta 288:401–409. (Available onine at: dx.doi.org/10.1007/s00425-008-0744-y) (verified 20 July 2011).
- Pelaz, S., G. S. Ditta, E. Baumann, E. Wisman, and M. F. Yanofsky. 2000. B and C floral organs identity function require SEPALLATA MADS-box genes. Nature 405:200-203. (Available online at: dx.doi.org/10.1038/35012103) (verified 20 July 2011).
- Vandenbussche, M., J. Zethof, E. Souer, R. Koes, G. B. Tomielli, M. Pezzotti, S. Ferrario, G. C. Angenent, and T. Gerats. 2003. Toward the analysis of the petunia MADS box gene family by reverse and forward transposon insertion mutagenesis approaches: B, C, and D foral organ identity functions require SEPALLATA-like MADS box genes in petunia. Plant Cell 15:2680–2693. (Available online at: http://dx.doi.org/10.1105/tpc.017376) (verified 20 July 2011).
Funding Statement
Development of this page was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, agreement 2009-85606-05673, administered by Michigan State University. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the United States Department of Agriculture.
PBGworks 1185
