ANOVA
Sample Size Estimation
Population Structure and Linkage
Association Analysis
Layout Specific Analyses
PBGworks 1557
PBGworks 1557
QGIS: A Free and Open Source Geographic Information System
HTP Geoprocessor: A plugin for QGIS
ASReml: Data analysis software designed for fitting linear mixed models
 Michael Gore is an associate professor of molecular breeding and genetics for nutritional quality at Cornell University in Ithaca, NY, where he is a member of the faculty in the Department of Plant Breeding and Genetics. He holds a BS and MS from Virginia Tech in Blacksburg, Virginia, and a PhD from Cornell University. Before joining the faculty at Cornell, he worked as a Research Geneticist with the USDA-ARS at the Arid-Land Agricultural Research Center in Maricopa, AZ. His expertise is in the field of quantitative genetics and genomics, especially the genetic dissection of metabolic traits. He has also contributed to the development and application of field-based, high-throughput phenotyping tools for plant breeding and genetics research. He teaches two short courses at the Tucson Winter Plant Breeding Institute in Tucson, Arizona, and serves on the editorial boards of Crop Science and Theoretical and Applied Genetics. His career accomplishments in plant breeding and genetics earned him the National Association of Plant Breeders Early Career Scientist Award in 2012 and the American Society of Plant Biologists Early Career Award in 2013.
Michael Gore is an associate professor of molecular breeding and genetics for nutritional quality at Cornell University in Ithaca, NY, where he is a member of the faculty in the Department of Plant Breeding and Genetics. He holds a BS and MS from Virginia Tech in Blacksburg, Virginia, and a PhD from Cornell University. Before joining the faculty at Cornell, he worked as a Research Geneticist with the USDA-ARS at the Arid-Land Agricultural Research Center in Maricopa, AZ. His expertise is in the field of quantitative genetics and genomics, especially the genetic dissection of metabolic traits. He has also contributed to the development and application of field-based, high-throughput phenotyping tools for plant breeding and genetics research. He teaches two short courses at the Tucson Winter Plant Breeding Institute in Tucson, Arizona, and serves on the editorial boards of Crop Science and Theoretical and Applied Genetics. His career accomplishments in plant breeding and genetics earned him the National Association of Plant Breeders Early Career Scientist Award in 2012 and the American Society of Plant Biologists Early Career Award in 2013.
 Kelly Thorp is a Research Agricultural Engineer with USDA-ARS in Maricopa, Arizona. He holds a BS and MS from the University of Illinois at Urbana-Champaign and a PhD from Iowa State University. His research focuses primarily on the development and application of informational technologies for monitoring cropping systems and understanding cropping system processes. Areas of expertise include remote sensing, cropping system simulation modeling, and geographic information systems. Application areas for these technologies include crop water and nitrogen status assessment, precision agriculture, management of nitrogen fertilizer, irrigation and drainage water management, field-based plant phenomics, and development of new bioenergy crops. He serves as an associate editor for Transactions of the ASABE and Applied Engineering in Agriculture.
Kelly Thorp is a Research Agricultural Engineer with USDA-ARS in Maricopa, Arizona. He holds a BS and MS from the University of Illinois at Urbana-Champaign and a PhD from Iowa State University. His research focuses primarily on the development and application of informational technologies for monitoring cropping systems and understanding cropping system processes. Areas of expertise include remote sensing, cropping system simulation modeling, and geographic information systems. Application areas for these technologies include crop water and nitrogen status assessment, precision agriculture, management of nitrogen fertilizer, irrigation and drainage water management, field-based plant phenomics, and development of new bioenergy crops. He serves as an associate editor for Transactions of the ASABE and Applied Engineering in Agriculture.
Barnett V, Lewis T (1994). Outliers in Statistical Data, 3rd edition, John Wiley, New York, NY, USA.
Belsley DA, Kuh E, Welsch RE (2004). Regression Diagnostics: Identifying Influential Data and Sources of Collinearity. Wiley-Interscience, Hoboken, NJ, USA.
Box GEP, Cox DR (1964). An analysis of Transformations. J. Roy. Stat. Soc. B. Met. 26: 211–252.
Cook RD, and Weisberg S (1982) Residuals and Influence in Regression, Chapman & Hall, New York, NY, USA
Henderson CR (2004). Applications of Linear Models in Animal Breeding. University of Guelph, Guelph, Ontario, Canada.
Holland JB, Nyguist WE, Cervantes-Martınez CT (2003). Estimating and interpreting heritability for plant breeding: an update. In Janick J (ed) Plant Breeding Reviews. John Wiley and Sons: Hoboken, New Jersey, USA.
Kutner MH, Nachtsheim CJ, Neter J, Li W (2004). Applied Linear Statistical Models, 4th ed. McGraw-Hill, Boston, MA, USA.
McCullagh P and Nelder JA (1989). Generalized Linear Models, Second Edition. Chapman & Hall, New York, NY, USA.
Development of this resource was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, and Dry Bean Root Health East Africa, Cotton Incorporated and United States Department of Agriculture – Agricultural Research Service (USDA-ARS). Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the United States Department of Agriculture. Mention of trade names or commercial products in this publication is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the USDA. The USDA is an equal opportunity provider and employer.
Field Phenomics Data Analysis Slides.pdf (6.36 MB)
PBGworks 1633
Original day/time: February 6, 2014 at 1:00pm Eastern Time (-05:00 GMT)
Databases
Field identification
Metagenomics
Find all upcoming and archived webinars »
Martin Chlivers is a visiting Assistant Professor at Michigan State University. He studies diseases of field crops, and the biology and genetics of fungal and oomycete organisms that cause disease. He utilizes both classic techniques such as culturing causal agents and the latest technologies, such as next generation sequencing of metagenomes, to improve disease management by understanding the organisms, factors, and host-pathogen interactions that drive disease.

PBGworks 1681
Allen Van Deynze
Univeristy of California Davis Seed Biotechnology Center
PBGworks 1483
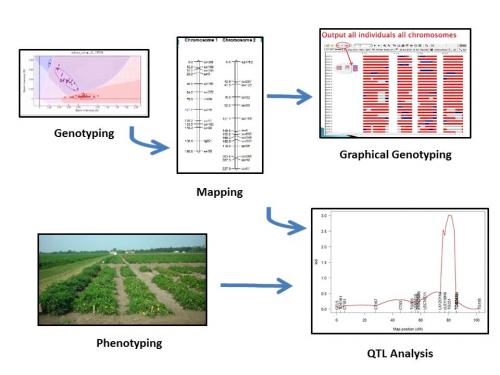
(Figure 1) Flow of information from collecting genotypic and phenotypic data to mapping to graphical genotyping and quantitative trait locus analysis. Figure credit: Heather Merk. Image credits: Genotyping, Allen Van Deynze, UC Davis; Mapping, Scott Wolfe, The Ohio State University; Graphical Genotyping, Nancy Huarachi, The Ohio State University; Phenotyping, David Francis, The Ohio State University; QTL Analysis, Hamid Ashrafi, UC Davis.
PBGworks 1563
Author:
Karolina Sikorska, Department of Biostatistics, Erasmus Medical Centre in Rotterdam
Part one explains GWA analysis in a loop using lm and lsfit functions and semi-parallel computations of linear regression with covariates. Also explains how to handle missing phenotype and SNP data.
Part two explains semi-parallel logisitic regression in R based on iteratively reweighted least squares (equivalent to glm), with and without covariates.
Part three explains how to convert the SNP matrix from a text file to an array-oriented binary file using the Ncdf and ff packages. Array-oriented binary files allow efficient access to blocks (columns) of SNPs by SNP, as opposed to by individual/line (rows).

Karolina Sikorska received a Master’s degree in Mathematics from the Gdansk University of Technology, Poland, with a specialization in financial mathematics. In 2009 she started her PhD project in the Department of Biostatistics, Erasmus Medical Centre in Rotterdam. Her research is related to fast computations in genome-wide association studies. Her work is focused on developing new methodology and algorithms which significantly speed up computations in GWAS for simple models, such as linear and logistic regression, as well as, mixed models for analyzing longitudinal data. She is also interested in improving tools for efficient data access in GWAS framework.
Development of this resource was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, Dry Bean Root Health East Africa, and the Erasmus Medical Center Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the United States Department of Agriculture.
Slides.pdf (570.88 KB)
PBGworks 1641

Nick Wheeler is a tree breeder with Molecular Tree Breeding Services, LLC, and is an adjunct professor at North Carolina State University. He has experience as a tree breeder for the British Columbia Ministry of Forests and as an industry scientist for Weyerhaeuser.
PBGworks 1688
PBGworks 1485
PBGworks 1564
 Duke Pauli is a graduate student at Montana State University. His work primarily focuses on the development of new malting barley varieties for the Montana production region using genome-wide association mapping to identify novel beneficial alleles that can be incorporated into barley breeding programs. His work also focuses on the implementation of genomic selection and understanding the utility of genomic selection on regional plant breeding programs.
Duke Pauli is a graduate student at Montana State University. His work primarily focuses on the development of new malting barley varieties for the Montana production region using genome-wide association mapping to identify novel beneficial alleles that can be incorporated into barley breeding programs. His work also focuses on the implementation of genomic selection and understanding the utility of genomic selection on regional plant breeding programs.
Photo Credit: Modified from Mina Talajoor
Development of this resource was supported in part by the National Institute of Food and Agriculture (NIFA) Solanaceae Coordinated Agricultural Project, Dry Bean Root Health East Africa, the Triticeae Coordinated Agriculture Project, and the Barley Coordinated Agriculture Project. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the United States Department of Agriculture.
GAPIT_Presentation_slides.pdf (19.4 MB)
Code for GAPIT webinar (1).R.zip (3.59 KB)
HapMap_genotypes.txt (5.29 MB)
protein.txt (14.22 KB)
PBGworks 1642